



版权说明:本文档由用户提供并上传,收益归属内容提供方,若内容存在侵权,请进行举报或认领
文档简介
1、Gene Manipulation&Genomic AnalysisRecombinant DNA cloning technologynnApplications of recombinant DNA technologynnGenomic analysisnnDNA sequencing1, Maxam-Gilbert sequencing2, Sanger-Coulson sequencing3, Next Generation SequencingMaxam-Gilbert sequencing (chemical cleavage method using double-st
2、randed (ds) DNA)1.Double-stranded DNA to be sequenced is labelled by attaching a radioactive phosphorus (32P) group to the 5' end.2.Using dimethyl sulphoxide and heating to 90oC, the two strands of the DNA are separated and purified3.Single-stranded sample is split into separate samples and each
3、 is treated with one of the cleavage reagents.4.If reactions have been arranged to give only one, or a few, cleavages per DNA molecule, a nested set of end-labelled DNA fragments of different lengths is produced.5.The samples are run together on a sequencing gel which separates the fragments by elec
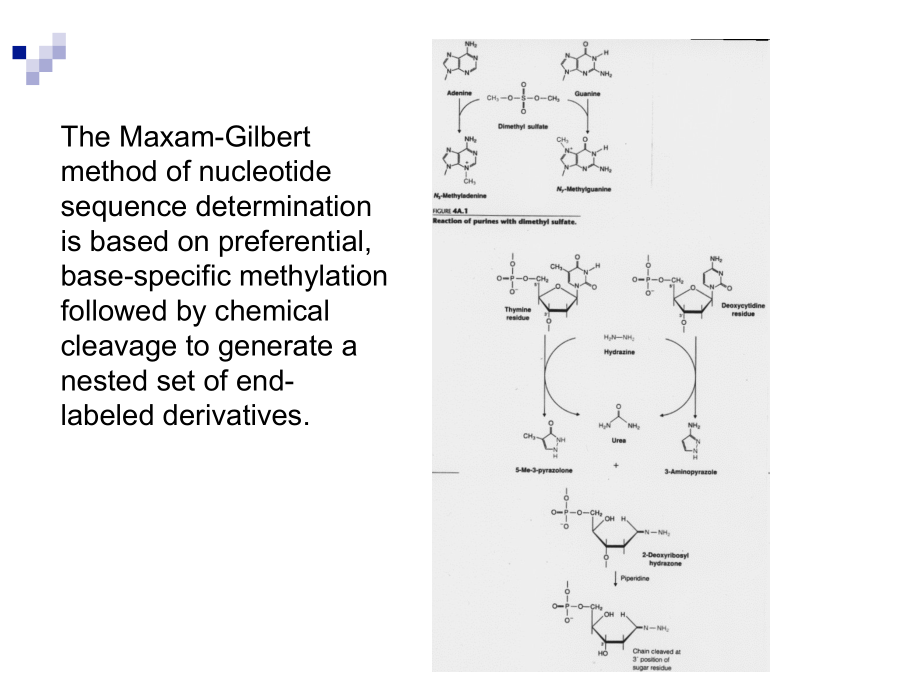
4、trophoresis depending on their size. DNA bands in the gel are visualized by autoradiography6.The DNA sequence is read directly from the gelThe Maxam-Gilbert method of nucleotide sequence determination is based on preferential, base-specific methylation followed by chemical cleavage to generate a nes
5、ted set of end- labeled derivatives.Maxam-Gilbert sequencingBase specificityChemical used for base alterationChemical used for altered base removalChemical used for strand cleavageGDimethylsulphatePiperidinePiperidineA+GAcidAcidPiperidineC+THydrazinePiperidinePiperidineCHydrazine + alkaliPiperidineP
6、iperidineA>CAlkaliPiperidinePiperidineMaxam-Gilbert sequencingSanger-Coulson sequencing (chain termination method using single-stranded (ss) DNA)1.Sample DNA to be sequenced is clonded into M13 vector DNA to generate ssDNA.2.A short oligonucleotide primer (usually chemically synthesized and somet
7、imes labelled ) is added to the ss recombinant DNA.3.DNA polymerase is then added in the presence of 4 normal nucleotides: d-ATP, d- CTP, d-GTP and d-TTP (one or more of which are labelled with 32P) and a low concentration of 4 analogues of the normal nucleotides in separate incubation mixes.4.Compl
8、ementary strand synthesis occurs away from the primer.5.Each of the 4 mixes is run together on a sequencing gel which separates the fragments by electrophoresis depending on their size. DNA bands in the gel are visualized by autoradiography.6.The DNA sequence is read directly from the gel in a simil
9、ar way to a Maxam-Gilbert sequencing gel.Structures of ribonucleoside triphosphate (NTP), deoxyribonucleoside triphosphate (dNTP), and dideoxyribonucleoside triphosphate (ddNTP)Sanger-Coulson sequencingSanger-Coulson sequencingDideoxy DNA sequencing of a theoretical DNA fragmentDideoxy Sequencing of
10、 DNAAUTOMATED DNA SEQUENCINGIt is based on the Sanger-Coulson chain termination method but the 4 different dideoxy nucleotides (ddA, ddC, ddG and ddT) are fluorescently labelled.4 different fluorophores are used, all 4 reactions can be run in the same tube.Genome SequencingChromosome WalkingGenome s
11、hotgun sequencingStrandOriginalSequenceAGCATGCTGCAGTCATGCTTAGGCTA AGCATGCTGCAGTCATGCT-TAGGCTAAGCATG-CTGCAGTCATGCTTAGGCTAAGCATGCTGCAGTCATGCTTAGGCTAFirst shotgun sequenceSecond shotgun sequenceReconstructionGenome shotgun sequencingHUMAN GENOME PROJECT (HGP)Hierarchical Shotgun (“map-based”, “BAC-base
12、d”, “clone-by-clone”)1990CELERA GENOMICS 1998Whole Genome ShotgunNext Generation Sequencing1. Roche 454 pyrosequencing2. Illumina (Solexa) sequencing3. SOLiD sequencingRoche 454 Sequencing,based on sequencing-by-synthesisNucleotides are flowed sequentially in a fixed order across the PicoTiterPlate
13、device during a sequencing run.During the nucleotide flow, hundreds of thousands of beads each carrying millions of copies of a unique single-stranded DNA molecule are sequenced in parallel.If a nucleotide complementary to the template strand is flowed into a well, the polymerase extends the existin
14、g DNA strand by adding nucelotide(s).Addition of one (or more) nucleotide(s) results in a reaction that generates a light signal that is recorded by the CCD camera in the instrument.The signal strength is proportional to the number of nucleotides incorporated in a single nucelotide flow.ssDNA templa
15、te is hybridized to a sequencing primer and incubated with the enzymes DNA polymerase, ATP sulfurylase, luciferase and apyrase, and with the substrates adenosine 5´ phosphosulfate (APS) and luciferin.Addition of one of the four dNTPs, DNA polymerase incorporates the correct, complementary dNTPs
16、 onto the template. This incorporation releases pyrophosphate (PPi) stoichiometrically.ATP sulfurylase quantitatively converts PPi to ATP in the presence of adenosine 5´ phosphosulfate.This ATP acts as fuel to the luciferase- mediated conversion of luciferin to oxyluciferin that generates visib
17、le light in amounts that are proportional to the amount of ATP. The light produced in the luciferase- catalyzed reaction is detected by a camera.Unincorporated nucleotides and ATP are degraded by the apyrase, and the reaction can restart with another nucleotide.Roche 454 SequencingIllumina SolexaIll
18、umina SolexaIllumina SolexaIllumina SolexaIllumina SolexaLife/APGs SOLiDThird Generation Sequencing第三代是单技术是为了解决第二的缺点而开发的,它的根本特点,不需要任何PCR的过程,这是为了能有效避免因PCR偏向性而导致的系统错误,同时提高读长,并要保持二代技术的高通量,低成 本的优点。1.基于电信号纳米单技术。当DNA碱基通过纳米,它们使电荷发生变化,从而短暂地影响流过纳米孔的电流强度(每种碱基所影响的电流变化幅度是不同的),灵敏的电子过的碱基。2.PacBio SMRT技术。在SMRTCell
19、(单检测到这些变化从而鉴定所通实时反应孔)中有许多圆形纳米小孔, 即 ZMW(零模波导孔),外径 100多纳米,比检测激光波长小(数百纳米),激光从底部打上去后不能穿透小孔进入上方溶液区,能量被限制在一个 小范围(体积20X 10-21 L)里,正好足够覆盖需要检测的部分,使得信号仅来自这个小反应区域, 景降到最低。中,从而实现将背过多游离核苷酸单体依然留在3.Ion Torrent。该技术使用了一种布满小孔的高密度半导体, 一个小反应池。当DNA聚合酶把核苷酸聚合到延伸中的DNA链上孔就是一个出一个氢离子,反应池中的PH发生改变,位于池下的离子感受时,会器感受到H+离子信号,H+离子信号再直
20、接转化为数字信号,从而读出DNA序列。Polymerase Chain Reaction (PCR)polymerase chain reaction (PCR)polymerase chain reaction (PCR)polymerase chain reaction (PCR)Diagnostic Applications of PCR detecting pathogens using genome-specific primer pairs screening specific genes for unknown mutations genotyping using known S
21、TS markersSubcloning DNA targets using PCR T/A Cloning Restriction Site Addition Blunt-end LigationPCR-mediated in vitro mutagenesisGeneration of DNA probesAmplification of differentially-expressed gene sequences Differential display reverse transcriptase PCR (DDRT-PCR) Suppression subtraction hybri
22、dization (SSH) Amplification of cell-specific transcripts using RT-PCRThe use of PCR in forensic science.Subcloning DNA targets using PCRNNN-EcoRIEcoRI-NNNEcoRI digestionClone to EcoRI sitePCR-mediated mutagenesis AATTCCP1 AAP3 AATTTTGGP2P4P1CCGGP4 CCGGPCR-mediated deletionP1P3P2P4P1P4Single strand
23、PCR to generate a DNA probePrimer 1 : Primer 2 = 10 : 1Primer 1Primer 2PCR to generate a template for in vitro transcriptionT7 promoterT7 polymeraseTo apply PCR to the study of RNA, the RNA sample mustfirst be reverse transcribed to cDNA to provide the necessary DNA template for the thermostable pol
24、ymerase. This process is called reverse transcription (RT), hence the name RT-PCR.Schematic diagram of two 5-RACE methods.Schematic diagram of a typical 3-RACE protocol.Quantitative Real-Time PCRQuantitative Real-Time PCRb TaqMan probeTaqman-based SNP genotypingQuantitative Real-Time PCRa representa
25、tive amplification plotQuantitative Real-Time PCRHigh-throughput droplet PCRssDNA template is hybridized to a sequencing primer and incubated with the enzymes DNA polymerase, ATP sulfurylase, luciferase and apyrase, and with the substrates adenosine 5´ phosphosulfate (APS) and luciferin.Analysis of Biological Processe
温馨提示
- 1. 本站所有资源如无特殊说明,都需要本地电脑安装OFFICE2007和PDF阅读器。图纸软件为CAD,CAXA,PROE,UG,SolidWorks等.压缩文件请下载最新的WinRAR软件解压。
- 2. 本站的文档不包含任何第三方提供的附件图纸等,如果需要附件,请联系上传者。文件的所有权益归上传用户所有。
- 3. 本站RAR压缩包中若带图纸,网页内容里面会有图纸预览,若没有图纸预览就没有图纸。
- 4. 未经权益所有人同意不得将文件中的内容挪作商业或盈利用途。
- 5. 人人文库网仅提供信息存储空间,仅对用户上传内容的表现方式做保护处理,对用户上传分享的文档内容本身不做任何修改或编辑,并不能对任何下载内容负责。
- 6. 下载文件中如有侵权或不适当内容,请与我们联系,我们立即纠正。
- 7. 本站不保证下载资源的准确性、安全性和完整性, 同时也不承担用户因使用这些下载资源对自己和他人造成任何形式的伤害或损失。
最新文档
- 2026年部编版本二年级下册《18.大象的耳朵》教案设计
- 餐厅食品安全培训课件
- 2026校招:鞍钢集团题库及答案
- 2026小学教师招聘真题及答案
- 2026中考冲刺动员大会校长发言稿:百日逐梦我们与你并肩前行
- 2026中考冲刺动员大会教师发言稿:百日奋进我们与你共谱辉煌
- 2025年中国旅游地理知识竞赛试卷及答案
- 船舶火灾应急演练总结报告范文
- 项目部安全生产责任制考核制度及考核表
- 2026年软件测试技术知识竞赛题库及答案
- 承德市市直医疗卫生单位招聘考试真题2024
- 2025年健身行业营销组合模式可行性分析报告
- 2025年固体废物分类处理环保治理计划书
- 金沙金坪山泉水厂建设项目入河排污口设置论证报告
- 衡阳市社区干部管理办法
- 2024新版 外研社版(三年级起点)三年级上册单词课本同步字帖
- 《最后的问题》和《终极答案》阿西莫夫
- 江南大学《高等数学Ⅱ(2)》2022-2023学年第一学期期末试卷
- 盖房四邻签字协议书范文
- 高一英语阅读理解试题(生活类)
- 农民工欠薪起诉书模板

评论
0/150
提交评论